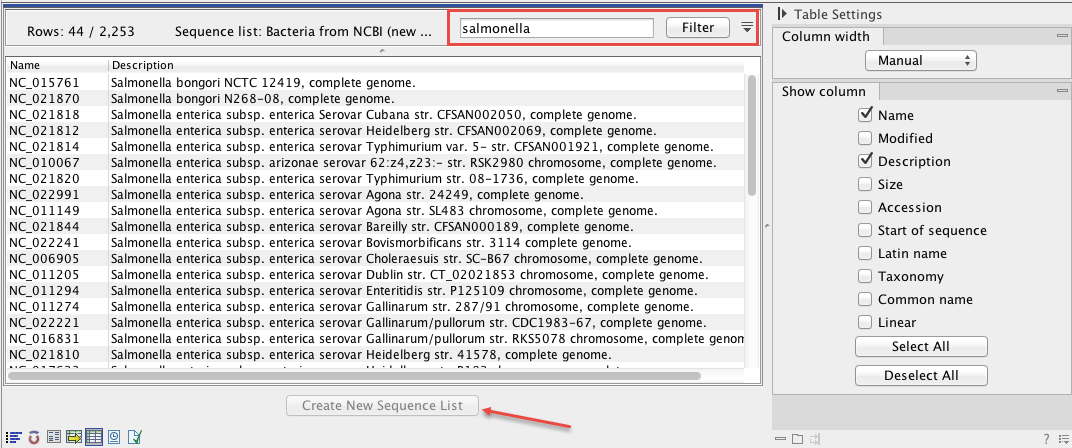
Figure 15.2: The downloaded NCBI bacterial genomes database was filtered for Salmonella data. A subset of 44 out of 2,253 sequences matched this search criterion.
After download, it is possible to select a subset of sequences and saving the reduced list in a new sequence list. This can reduce subsequent analysis runtime significantly.
For example, from a collection of bacterial genomes that include multiple representatives of each genus, you can extract a genus specific subset of sequences to a new list:

Figure 15.2: The downloaded NCBI bacterial genomes database was filtered for Salmonella data. A subset of 44 out of 2,253 sequences matched this search criterion.
Another way to extract a subset of a database is to make use of the Split Sequence List tool. For more information, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Split_Sequence_List.html.