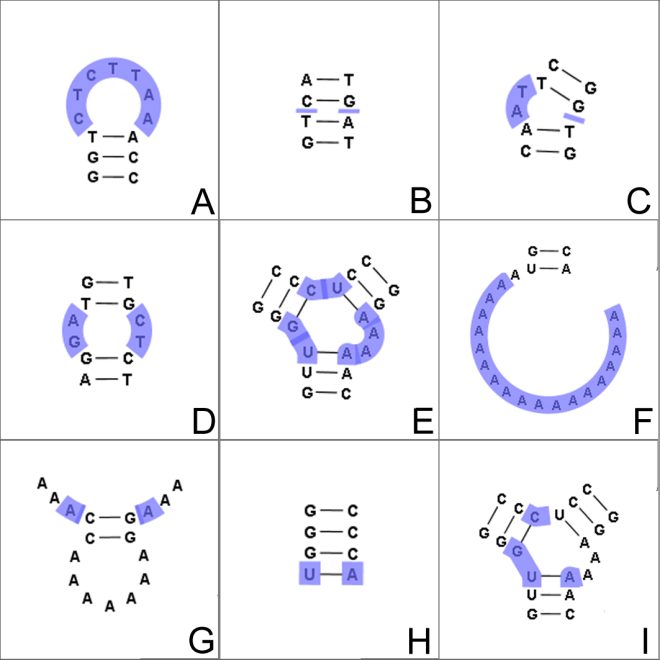
Figure 26.29: The different structure elements of RNA secondary structures predicted with the free energy minimization algorithm in CLC Genomics Workbench. See text for a detailed description.
In this section, we classify the structure elements defining a secondary structure and describe their energy contribution.

Figure 26.29: The different structure elements of RNA secondary structures predicted with the free energy minimization algorithm in CLC Genomics Workbench. See text for a detailed description.
The structure elements involving nested base pairs can be classified by a given base pair and the other base pairs that are nested and accessible from this pair. For a more elaborate description we refer the reader to [Sankoff et al., 1983] and [Zuker and Sankoff, 1984].
If the nucleotides with position number ![]() form a base pair
and
form a base pair
and
![]() , then we say that the base pair
, then we say that the base pair ![]() is
accessible from
is
accessible from ![]() if there is no intermediate base pair
if there is no intermediate base pair
![]() such that
such that
![]() . This means that
. This means that ![]() is nested within the pair
is nested within the pair ![]() and there is no other base pair
in between.
and there is no other base pair
in between.
Using the number of accessible pase pairs, we can define the following distinct structure elements:
A number of techniques are available for probing RNA structures. These techniques can determine individual components of an existing structure such as the existence of a given base pair. It is possible to add such experimental constraints to the secondary structure prediction based on free energy minimization (see figure 26.30) and it has been shown that this can dramatically increase the fidelity of the secondary structure prediction [Mathews and Turner, 2006].
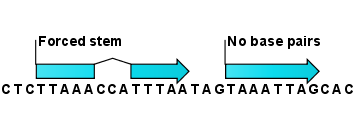
Figure 26.30: Known structural features can be added as constraints to the secondary structure prediction algorithm in CLC Genomics Workbench.