CLC Genomics Workbench offers advanced and versatile options to create lists of sequence patterns or known motifs, represented either by a literal string or a regular expression.
A motif list can be created using one of two ways:
Toolbox | Classical Sequence Analysis (![]() ) | General Sequence Analysis (
) | General Sequence Analysis (![]() )| Create Motif List (
)| Create Motif List (![]() )
)
File | New | Motif List (![]() )
)
Add (![]() ) button at the bottom of the view. This will open a dialog shown in figure 14.32.
) button at the bottom of the view. This will open a dialog shown in figure 14.32.
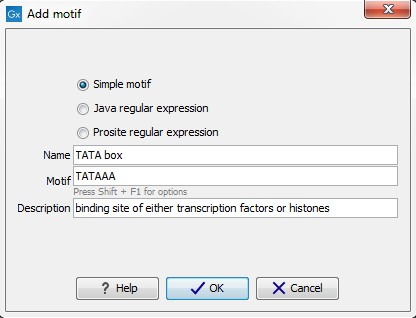
Figure 14.32: Entering a new motif in the list.
In this dialog, you can enter the following information:
The motif list can contain a mix of different types of motifs. This is practical because some motifs can be described with the simple syntax, whereas others need the more advanced regular expression syntax.
Instead of manually adding motifs, you can Import From Fasta File (![]() ). This will show a dialog where you can select a fasta file on your computer and use this to create motifs. This will automatically take the name, description and sequence information from the fasta file, and put it into the motif list. The motif type will be "simple".
). This will show a dialog where you can select a fasta file on your computer and use this to create motifs. This will automatically take the name, description and sequence information from the fasta file, and put it into the motif list. The motif type will be "simple".
Besides adding new motifs, you can also edit and delete existing motifs in the list. To edit a motif, either double-click the motif in the list, or select and click the Edit (![]() ) button at the bottom of the view.
) button at the bottom of the view.
To delete a motif, select it and press the Delete key on the keyboard. Alternatively, click Delete (![]() ) in the Tool bar.
) in the Tool bar.
Save the motif list in the Navigation Area, and you will be able to use for Motif Search (![]() ).
).