The dialog box contains three fields:
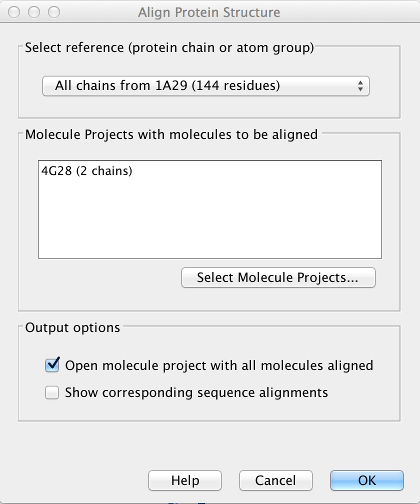
Figure 13.15: The Align Protein Structure dialog box.
- Select reference (protein chain or atom group) This drop-down menu shows all the protein chains and residue-containing atom groups in the current Molecule Project. Choosing from this menu tells the structure aligner to improve the quality of the alignment in this region at the cost of reducing the quality of the alignment elsewhere. The 'All chains from Molecule Project option will create a global alignment to all protein chains in the project, fitting e.g. a dimer to a dimer.
- Molecule Projects with molecules to be aligned One or more Molecule Projects containing protein chains may be selected.
- Output options The default output is a single Molecule Project containing all the input projects rotated onto the coordinate system of the reference. Several alignment statistics, including the RMSD, TM-score, and sequence identity, are added to the History of the output Molecule Project. Additionally, a sequence alignment between each project and the reference may be output.

