BLAST against local data
Running BLAST searches on your local machine can have several advantages over running the searches remotely at the NCBI:
- It can be faster.
- It does not rely on having a stable internet connection.
- It does not depend on the availability of the NCBI BLAST blast servers.
- You can use longer query sequences.
- You use your own data sets to search against.
On a technical level, the CLC Genomics Workbench uses the NCBI's blast+ software (see ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). Thus, the results of using a particular data set to search the same database, with the same search parameters, would give the same results, whether run locally or at the NCBI.
There are a number of options for what you can search against:
To conduct a BLAST search:
or Toolbox | BLAST ( ) | Local BLAST (
) | Local BLAST ( )
)
This opens the dialog seen in figure 12.5:
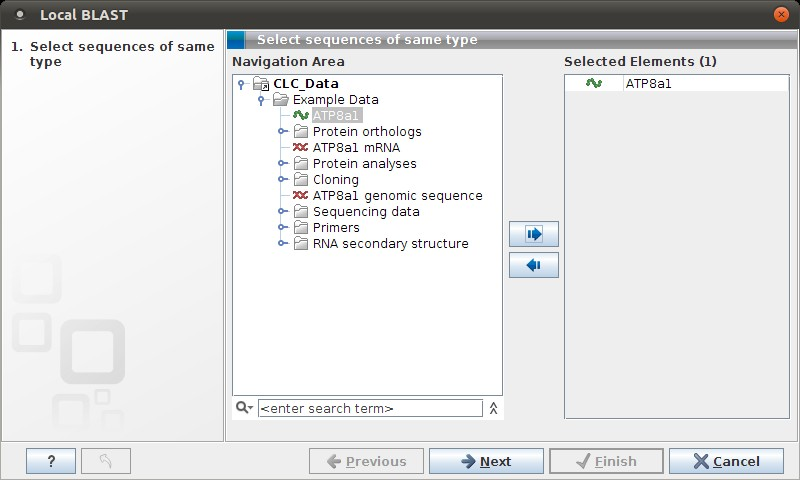
Figure 12.5: Choose one or more sequences to conduct a BLAST search.
Select one or more sequences of the same type (DNA or protein) and click Next.
This opens the dialog seen in figure 12.6:
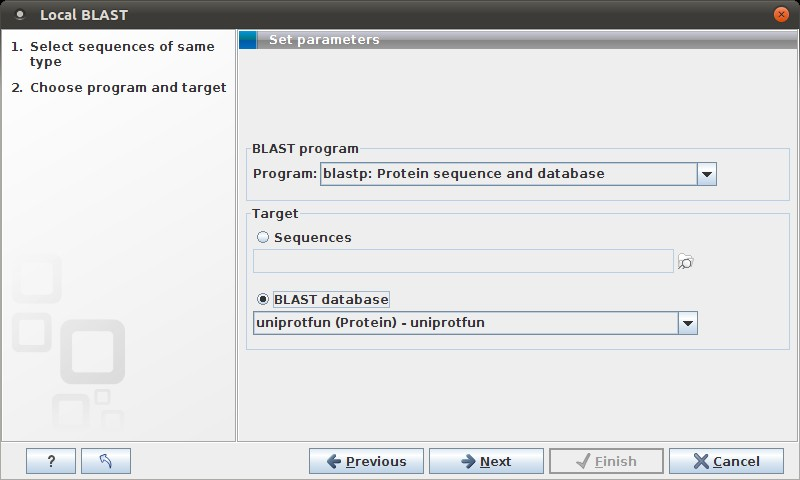
Figure 12.6: Choose a BLAST program and a target database.
At the top, you can choose between different BLAST programs. See
NCBI BLAST
for information about these methods.
You then specify the target database to use:
- Sequences. When you choose this option, you can use sequence data from the Navigation Area as database by clicking the Browse and select icon (
 ). A temporary BLAST database will be created from these sequences and used for the BLAST search. It is deleted afterwards. If you want to be able to click in the BLAST result to retrieve the hit sequences from the BLAST database at a later point, you should not use this option; create a create a BLAST database first.
). A temporary BLAST database will be created from these sequences and used for the BLAST search. It is deleted afterwards. If you want to be able to click in the BLAST result to retrieve the hit sequences from the BLAST database at a later point, you should not use this option; create a create a BLAST database first.
- BLAST Database. Select a database already available in one of your designated BLAST database folders. Read more in Manage BLAST databases.
When a database or a set of sequences has been selected, click Next.
This opens the dialog seen in figure 12.7:
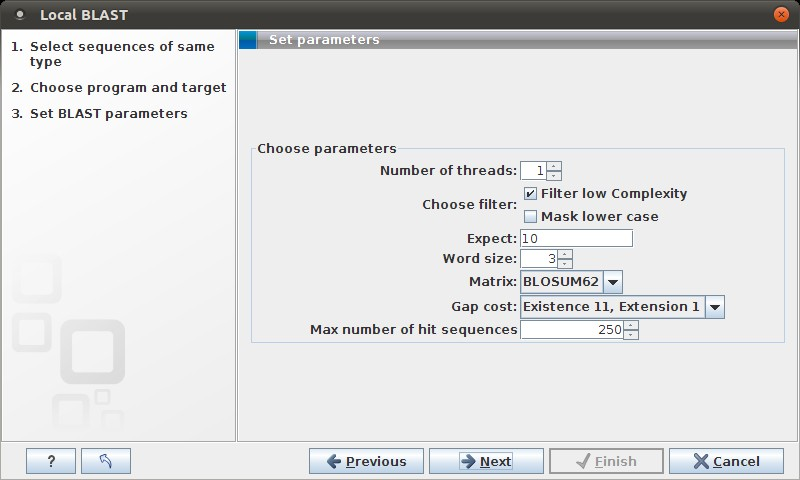
Figure 12.7: Examples of parameters that can be set before submitting a BLAST search.
See NCBI BLAST for information about these limitations.
There is one setting available for local BLAST jobs that is not relevant for remote searches at the NCBI:
- Number of processors. You can specify the number of processors which should be used if your Workbench is installed on a multi-processor system.
![]() ) | Local BLAST (
) | Local BLAST (![]() )
)


