How to run the Filter Causal Variants (WES-HD) workflow
To run the Filter Causal Variants (WES-HD) workflow, go to:
Toolbox | Ready-to-Use Workflows | Whole Exome Sequencing (![]() ) | Hereditary Disease (
) | Hereditary Disease (![]() ) | Filter Causal Variants (WES -HD) (
) | Filter Causal Variants (WES -HD) (![]() )
)
- Double-click on the Filter Somatic Variants (WES-HD) tool to start the analysis. If you are connected to a server, you will first be asked where you would like to run the analysis.
- Select the variant track you want to use for filtering causal variants (figure 13.48).
The panel in the left side of the wizard shows the kind of input that should be provided. Select by double-clicking on the variant track name or click once on the file and then click on the arrow pointing to the right side in the middle of the wizard.
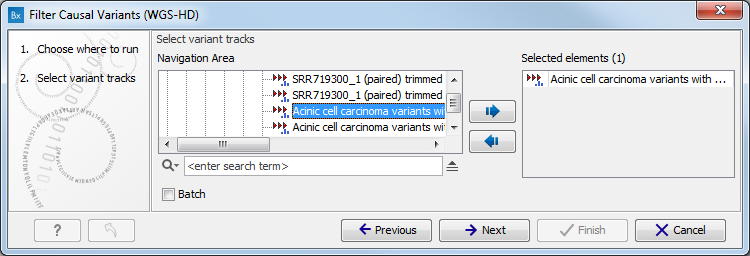
Figure 13.48: Select the variant track from which you would like to filter somatic variants. - Specify which of the 1000 Genomes populations should be used for annotation (figure 13.49).
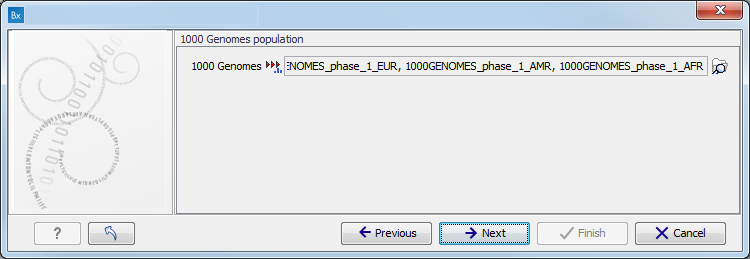
Figure 13.49: Select the relevant 1000 Genomes population(s). - Specify the 1000 Genomes population that should be used for filtering out variants found in the 1000 Genomes project.
This can be done using the drop-down list found in this wizard step. Please note that the populations available from the drop-down list can be specified with the Data Management (
 ) function found in the top right corner of the Workbench (see Download and configure reference data).
) function found in the top right corner of the Workbench (see Download and configure reference data).
- Specify the Hapmap populations that should be used for filtering out variants found in Hapmap (figure 13.50).
This can be done using the drop-down list found in this wizard step. Please note that the populations available from the drop-down list can be specified with the Data Management (
 ) function found in the top right corner of the Workbench (see Download and configure reference data).
) function found in the top right corner of the Workbench (see Download and configure reference data).
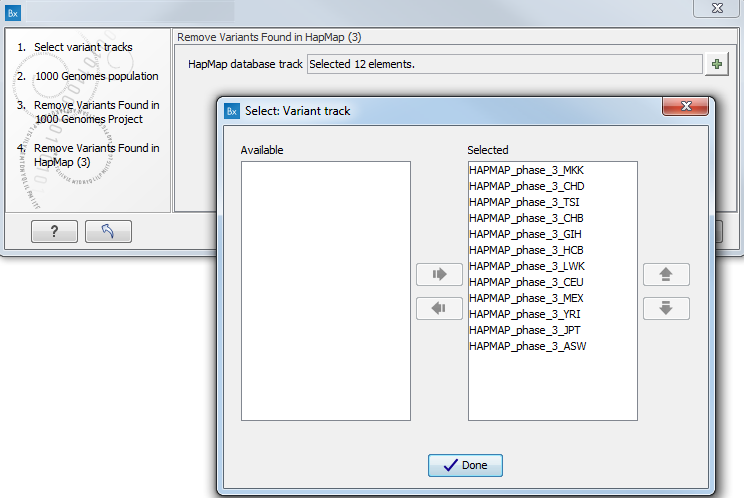
Figure 13.50: Select the relevant Hapmap population(s). - Pressing the button Preview All Parameters allows you to preview all parameters. At this step you can only view the parameters, it is not possible to make any changes. Choose to save the results and click on the button labeled Finish.
