Read statistics
This section contains simple statistics for all mapped reads, non-specific matches (reads that match more than place during the assembly), non-perfect matches and paired reads. Note! Paired reads are counted as two, even though they form one pair. The section on paired reads also includes information about paired distance and counts the number of pairs that were broken due to:
- Wrong distance
- When starting the mapping, a distance interval is specified. If the reads during the mapping are placed outside this interval, they will be counted here.
- Mate inverted
- If one of the reads has been matched as reverse complement, the pair will be broken (note that the pairwise orientation of the reads is determined during import).
- Mate on other contig
- If the reads are placed on different contigs, the pair will also be broken.
- Mate not matched
- If only one of the reads match, the pair will be broken as well.
Below these tables follow two graphs showing distribution of paired distances (see figure 19.18) and distribution of read lengths. Note that the distance includes both the read sequence and the insert between them as explained in General notes on handling paired data.
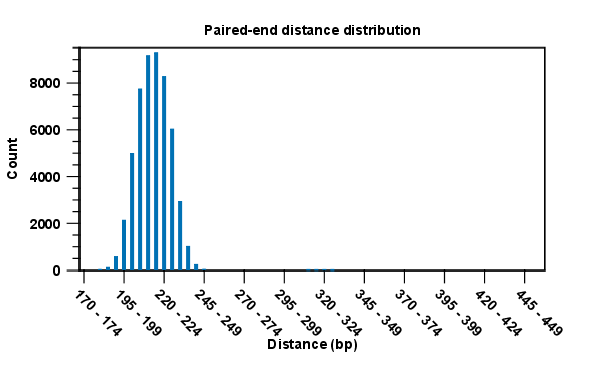
Figure 19.7: A bar plot showing the distribution of distances between intact pairs.
Two plots of the distribution of insertion and deletion lengths can bee seen in figure 19.19 and figure 19.20.
