Amino acid changes
This tool annotates variants with amino acid changes given a track with coding regions and a reference sequence (see figure 26.37).
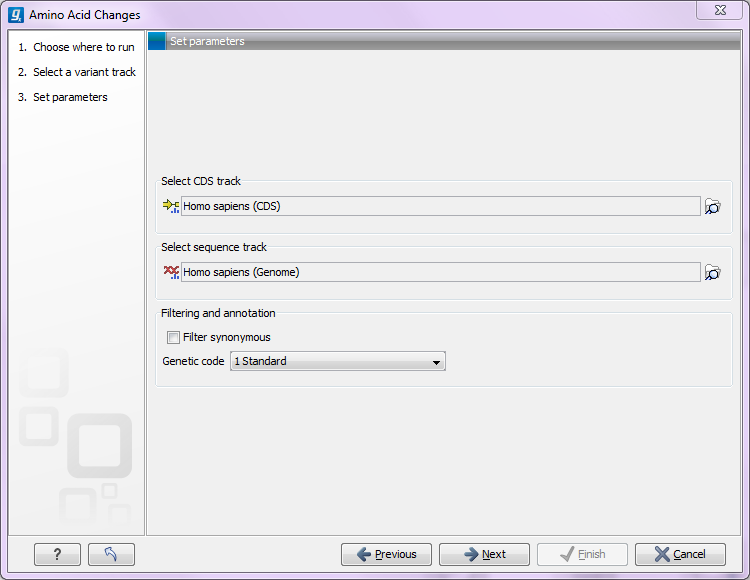
Figure 26.37: The amino acid changes annotation tool.
The result is a new track where each variant has information about the effect on the amino acid sequence of the corresponding protein. By filtering in the table view of this track on the column "Non-synonymous" for "Yes" only variants that change the protein product will be retained in the result track.
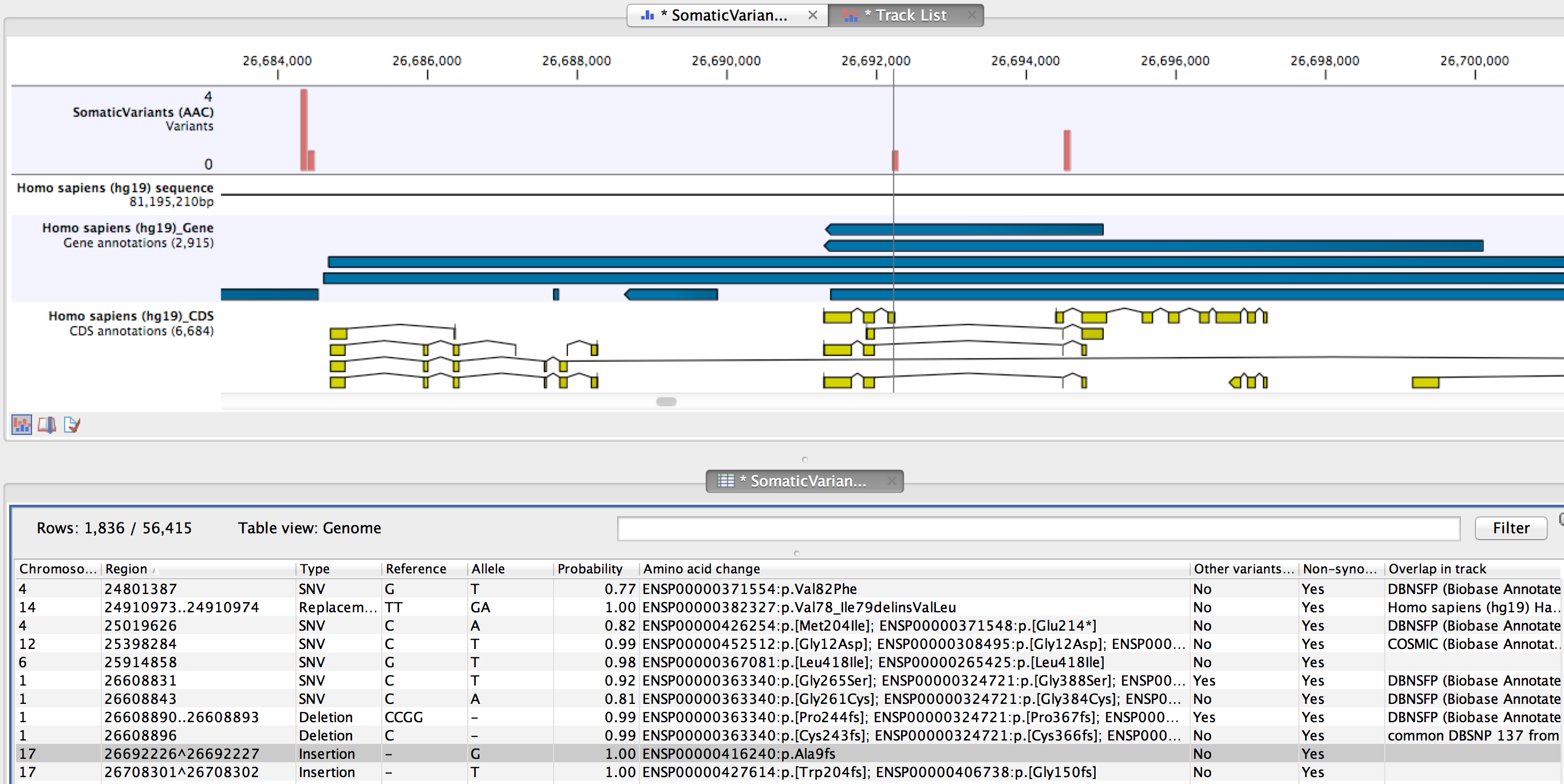
Figure 26.38: The resulting amino acid changes in track and table views.
An example of the output is given in Figure 26.38. The top track view displays the variant track, sequence track, gene annotation and CDS track. The lower table view is filtered for non-synonymous variants. Note that there is a column "Other variants within codon" set to "Yes" if other variants within the same codon are present that also could potentially cause amino acid changes. The column "Amino acid change" lists the effects on the protein sequence. For example, single amino-acid changes caused by SNVs are listed as "p.[Gly261Cys]", denoting that in the protein sequence (hence the "p.") the Glycine at position 261 is changed into Cysteine. Frame-shifts caused by indels are listed with the extension fs, for example p.[Pro244fs] denoting a frameshift at position 244 coding for Proline. For further details of the nomenclature see the "Recommendations for the description of protein sequence variants (v2.0)" at http://www.hgvs.org/mutnomen/.
