BLAST table
In addition to the
graphical display of a BLAST result, it is possible to view the
BLAST results in a tabular view. In the tabular view, one can get a
quick and fast overview of the results. Here you can also select
multiple sequences and download or open all of these in one single
step. Moreover, there is a link from each sequence to the sequence
at NCBI. These possibilities are either available through a
right-click with the mouse or by using the buttons below the table.
If the BLAST table view was not selected in Step 4 of the BLAST search, the table can be shown in the following way:
Click the Show BLAST Table button ( ) at the
bottom of the view
) at the
bottom of the view
Figure 12.10 is an example of a BLAST Table.
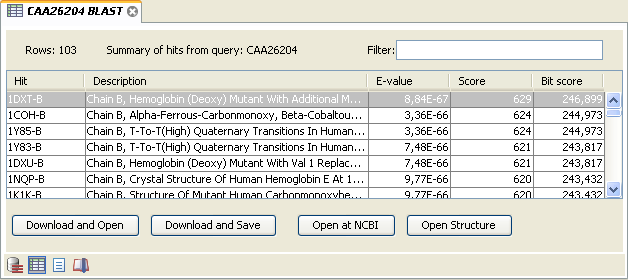
Figure 12.10: Display of the output of a BLAST search in the tabular view. The hits can be sorted by the different columns, simply by clicking the column heading.
The BLAST Table includes the following information:
- Query sequence. The sequence which was used for the search.
- Hit. The Name of the sequences found in the BLAST search.
- Id. GenBank ID.
- Description. Text from NCBI describing the sequence.
- E-value. Measure of quality of the match.
Higher E-values indicate that BLAST found a less homologous sequence.
- Score. This shows the score of the local alignment generated through the BLAST search.
- Bit score. This shows the bit score of the local alignment generated through the BLAST search. Bit scores are normalized, which means that the bit scores from different alignments can be compared, even if different scoring matrices have been used.
- Hit start. Shows the start position in the hit sequence
- Hit end. Shows the end position in the hit sequence.
- Hit length. The length of the hit.
- Query start. Shows the start position in the query sequence.
- Query end. Shows the end position in the query sequence.
- Overlap. Display a percentage value for the overlap of the query sequence and hit sequence. Only the length of the local alignment is taken into account and not the full length query sequence.
- Identity. Shows the number of identical residues in the query and hit sequence.
- %Identity. Shows the percentage of identical residues in the query and hit sequence.
- Positive. Shows the number of similar but not necessarily identical residues in the query and hit sequence.
- %Positive. Shows the percentage of similar but not necessarily identical residues in the query and hit sequence.
- Gaps. Shows the number of gaps in the query and hit sequence.
- %Gaps. Shows the percentage of gaps in the query and hit sequence.
- Query Frame/Strand. Shows the frame or strand of the query sequence.
- Hit Frame/Strand. Shows the frame or strand of the hit sequence.
In the BLAST table view you can handle the hit sequences. Select one or more sequences from the table, and apply one of the following functions.
- Download and Open.
Download the full sequence from NCBI and opens it. If multiple sequences are selected, they will all open (if the same sequence is listed several times, only one copy of the sequence is downloaded and opened).
- Download and Save.
Download the full sequence from NCBI and save it. When you click the button, there will be a save dialog letting you specify a folder to save the sequences. If multiple sequences are selected, they will all open (if the same sequence is listed several times, only one copy of the sequence is downloaded and opened).
- Open at NCBI.
Opens the corresponding sequence(s) at GenBank at NCBI.
Here is stored additional information regarding the selected
sequence(s). The default Internet browser is used for this purpose.
- Open structure. If the hit sequence contain structure information, the sequence is opened in a text view or a 3D view (3D view in CLC Main Workbench or CLC Genomics Workbench).
The hits can be sorted by the different columns, simply by clicking the column heading. In cases where individual rows have been selected in the table, the selected rows will still be selected after sorting the data.
You can do a text-based search in the
information in the BLAST table by using the filter at the upper
right part of the view. In this way you can search for e.g. species
or other information which is typically included in the
"Description" field.
The table is integrated with the graphical
view so that selecting a hit in the table will
make a selection on the corresponding sequence in the graphical
view.
![]() ) at the
bottom of the view
) at the
bottom of the view
