How to run the Correct PacBio Reads tool
To start the tool, go to:
Toolbox | Legacy Tools (![]() ) | Correct PacBio Reads (legacy) (
) | Correct PacBio Reads (legacy) (![]() )
In this dialog, you can select one or more sequence lists containing
the raw PacBio reads that should be corrected.
)
In this dialog, you can select one or more sequence lists containing
the raw PacBio reads that should be corrected.
Click Next to set the parameters for the error correction. This opens the dialog shown in figure 14.4.
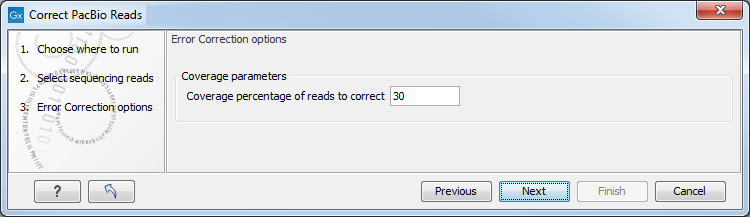
Figure 14.4: Set Coverage percentage of reads to correct for the error correction.
In this dialog, you can set the Coverage percentage of reads to correct. The error correction tool will correct a number of long reads amounting to the entered fraction of the total coverage. The remaining shorter reads are used to perform the correction. For example, if the Coverage percentage of reads to correct is set to 25%, the tool will correct a subset of the longest reads that amounts to 25% of the total coverage using the remaining shorter reads. The De Novo Assemble PacBio Reads (legacy) tool needs at least 25-30x coverage on microbial genomes in order to obtain a high-quality assembly. Thus, the Coverage percentage of reads to correct should be chosen such that the corrected reads supply a coverage of at least 25-30x. This means that if your dataset has coverage of about 200x, you should set Coverage percentage of reads to correct to 12-15%. For datasets with very high coverage, you can get a better error correction by lowering the Coverage percentage of reads to correct and at the same time get a sufficiently high coverage by the corrected reads to obtain a good assembly quality. Any reads containing ambiguous nucleotides are discarded before the Coverage percentage of reads to correct is calculated and will not be used in the error correction.
Click Next to set the output options, and click Finish to start the error correction.
