Setup Binding Site
Before a small molecule can be fitted to a protein binding pocket in a docking simulation, you must first specify where on the protein the binding site is found, to limit the search for the optimal protein-ligand complex. This is done using the Setup Binding Site action from the Molecule Project Side Panel. This action will open an interactive dialog box, and when the dialog box is closed with an "OK", a Binding Site Setup will appear in the Project Tree, and the Molecule Project is now ready to use for docking. The binding site setup can later be inspected and altered, by re-invoking the action from the Side Panel or by double-clicking the Binding Site Setup entry in the Project Tree (figure 10.45).
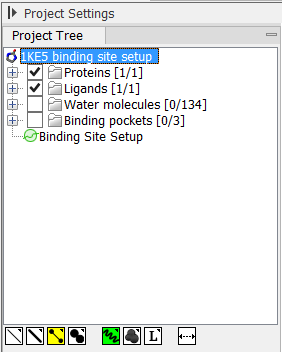
Figure 10.45: "Binding Site Setup" in the Project Tree.
In the following, each category in the Setup Binding Site dialog box will be described in detail. The 3D view will zoom to the different elements, when they are selected in the dialog box. This can be avoided by unchecking the Auto-zoom box in the lower left corner of the dialog box.
Binding site
The ligand binding mode search is carried out inside the binding site volume. It is specified using a sphere, which should be centered around the expected binding pocket, and have a radius large enough to accommodate all ligands docked to the protein. The binding site is shown as a transparent green sphere when invoking the Setup Binding Site action from the Molecule Project Side Panel (figure 10.46).
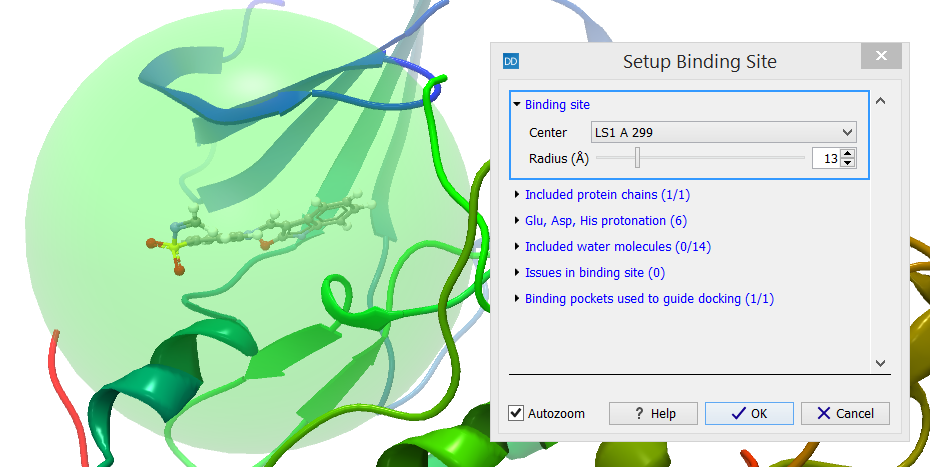
Figure 10.46: The binding site is shown as a transparent green sphere when invoking the Setup Binding Site action.
The center of the binding site can be selected from a list of the ligands, binding pockets and atom groups contained in the Molecule Project. Alternatively, the mouse can be used to select a number of atoms from the 3D view, e.g. residues known to be involved in binding using the 'Current atom selection' option. The radius of the sphere can also be adjusted. The sphere should be as small as possible, to improve the docking search efficiency, but large enough to ensure that proper ligand binding modes will not be discarded due to atoms not being able to fit inside the sphere.
Included protein chains (X/Y)
The category header indicates the number of protein chains that are included in the binding site setup (X) as well as the number of protein chains that have atoms inside the binding site volume (Y) (figure 10.47).
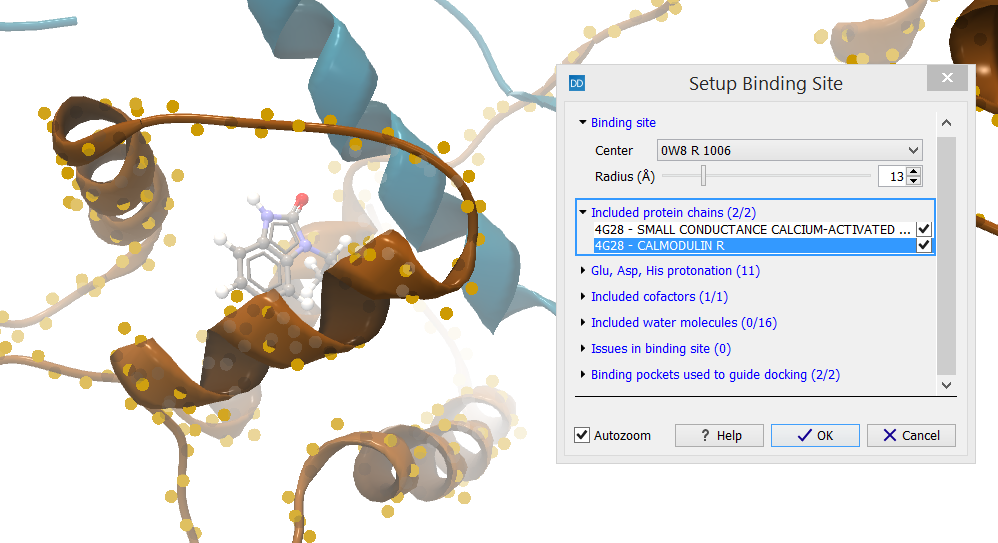
Figure 10.47: In the category header is indicated how many protein chains are selected to be part of the binding site setup (X) and how many protein chains have atoms inside the binding site volume (Y).
The protein chains found inside the binding site volume are listed. Selecting a row in the list will fit the view around the protein chain and highlight it. All protein chains found inside the binding site volume are included per default, but individual chains can be removed from the setup by unchecking the boxes next to them in the dialog box. This will not remove them from the Molecule Project, only from the binding site setup.
Glu, Asp, His protonation (X)
The "Glu, Asp, His protonation" category header indicates how many Glu, Asp, and His residues (X) that are found inside the binding site volume on the included protein chains. These residues are interesting as the protonation state of the side chain depends on the local environment. Per default, Glu and Asp (pKa~4) are setup with no protons on the acid group, and His (pKa~6) with a single proton positioned at N![]() (figure 10.48).
(figure 10.48).
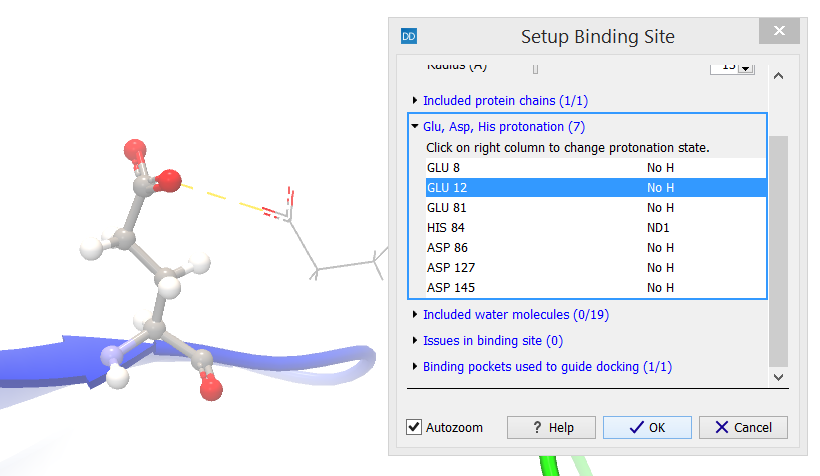
Figure 10.48: The protonation state is shown in the right-most column.
The residues are listed, and selecting an entry in the list will zoom to that residue and show it in a ball-and-sticks representation. The protonation of that residue can then be changed by clicking on the right-most column, which will make a list of protonation choices appear. Selecting an alternative protonation will immediately update the view (figure 10.49). If the selected side chain can form hydrogen bonds, these are automatically shown with blue dashed lines. Hydrogen bonds that could be formed if the protonation was changed are shown with yellow dashed lines.
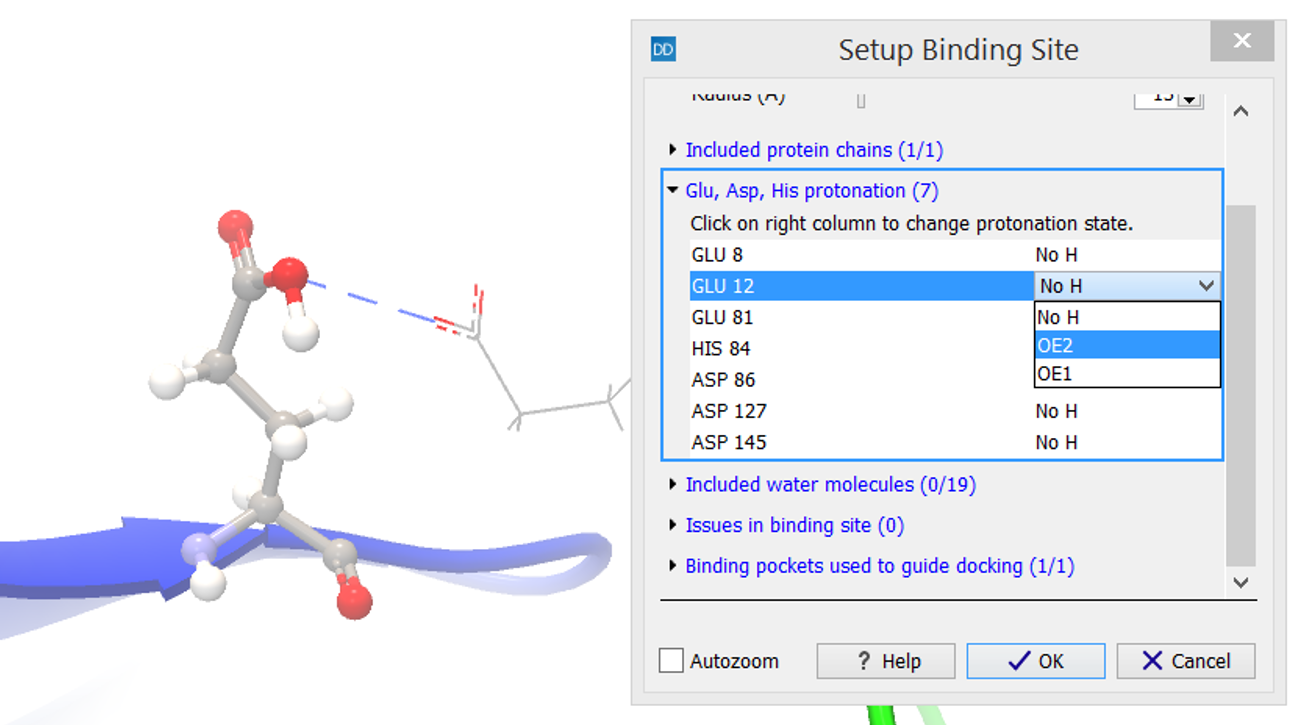
Figure 10.49: The protonation of a residue can be changed by clicking on the right-most column, which will make a list of protonation choices appear.
You can see more residues at the same time by selecting multiple entries from the list.
Note! Changing the protonation of residues will take immediate effect and will not revert if you press "Cancel" on the Setup Binding Site dialog box. However, like all other changes to molecules, it can be reverted using the "Undo" button.
Included cofactors (X/Y)
The category header indicates the number of cofactors that are included in the binding site setup (X) as well as the number of cofactors that have atoms inside the binding site volume (Y) (figure 10.50).
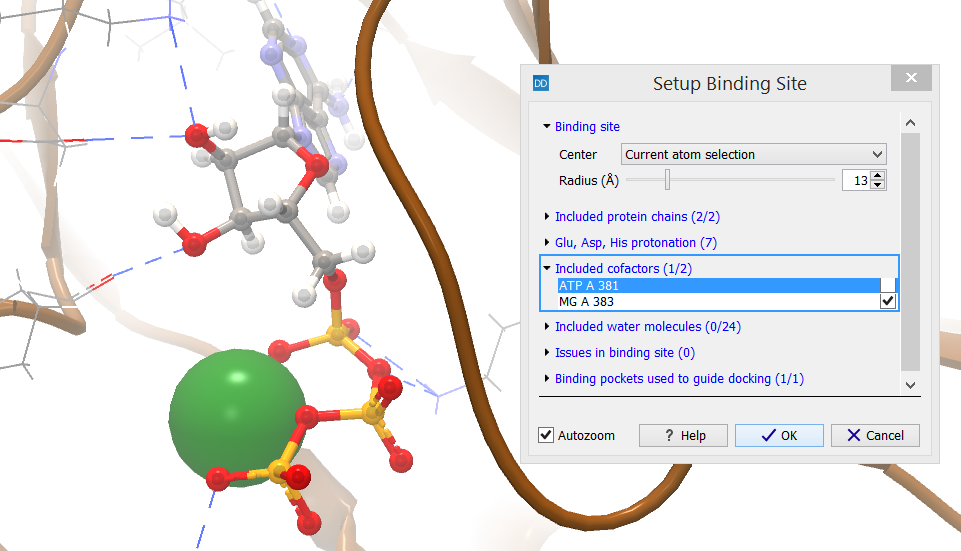
Figure 10.50: Cofactors within the binding site volume are included in the setup per default.
The cofactors found inside the binding site are listed. Selecting a row in the list will zoom the view around it and high-light it. All cofactors found inside the binding site are included per default, but individual cofactors can be removed from the setup by unchecking the boxes next to them in the dialog box. This will not remove them from the Molecule Project, only from the binding site setup.
Included water molecules (X/Y)
The category header indicates the number of water molecules that are included in the binding site setup (X) as well as the number of water molecules that are found inside the binding site volume (Y) (figure 10.51).
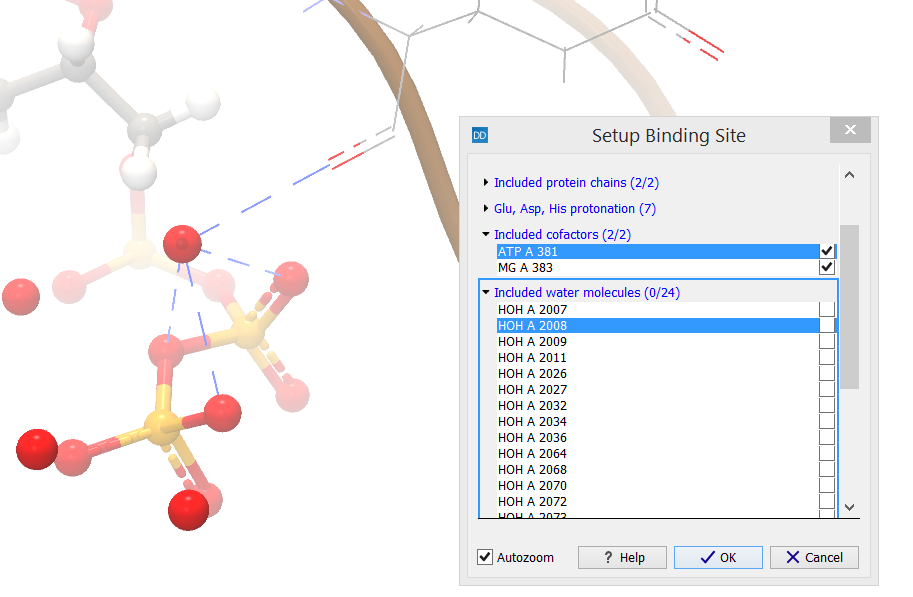
Figure 10.51: The category header indicates the number of water molecules that are included in the binding site setup (X) as well as the number of water molecules that are found inside the binding site volume (Y).
The water molecules found inside the binding site are listed. Selecting one or more rows in the list will zoom the view around the selected water molecules and high-light them. If the water molecules are currently not displayed, their position will be seen as brown atom-selection dots. Hydrogen bond interactions involving the selected water molecules and other molecules included in the binding site setup will automatically be shown. Per default, water molecules are not included in the binding site setup, but individual molecules can be included by checking the boxes next to them in the dialog box.
Note As all molecules included in the setup will stay rigid and take up space in the binding pocket during docking, only include water molecules that you know is important for binding.
Included nucleic acids (X/Y)
The category header indicates the number of nucleic acid chains that are included in the binding site setup (X) and how many nucleic acid chains have atoms inside the binding site volume (Y) (figure 10.52).
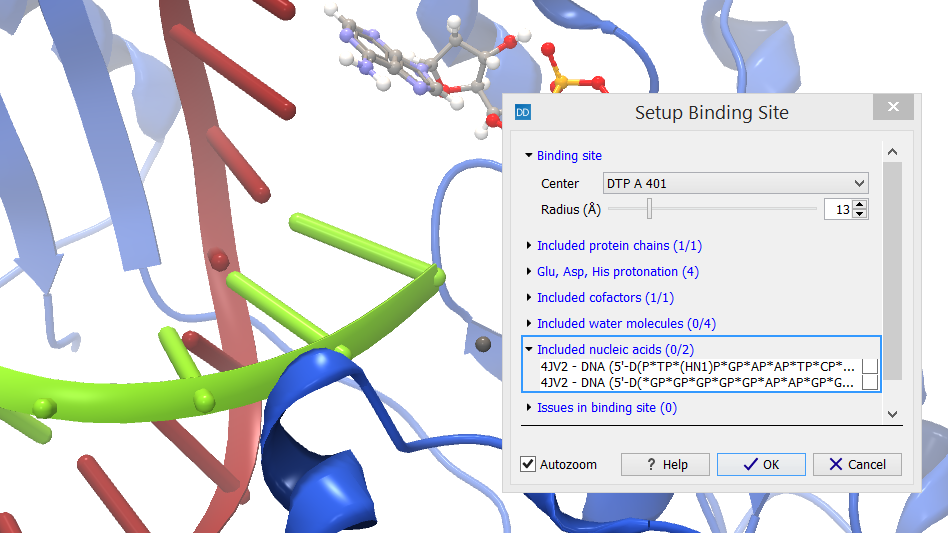
Figure 10.52: Select nucleic acid chains to include in the target.
The nucleic acids found inside the binding site are listed. Selecting a row in the list will zoom the view around the chain and high-light it. All nucleic acids found inside the binding site volume are included per default, but individual chains can be removed from the setup by un-checking the boxes next to them in the dialog box.
Note The docking score has not been optimized to dock ligands against nucleic acids, and it is not advisable to have nucleic acids as the main target for the docking.
Issues in binding site (X)
The category header indicates the number (X) of the issues in the Issues list (see Import issues) that relates to atoms or molecules found inside the binding site volume and included in the Binding Site Setup (figure 10.53).
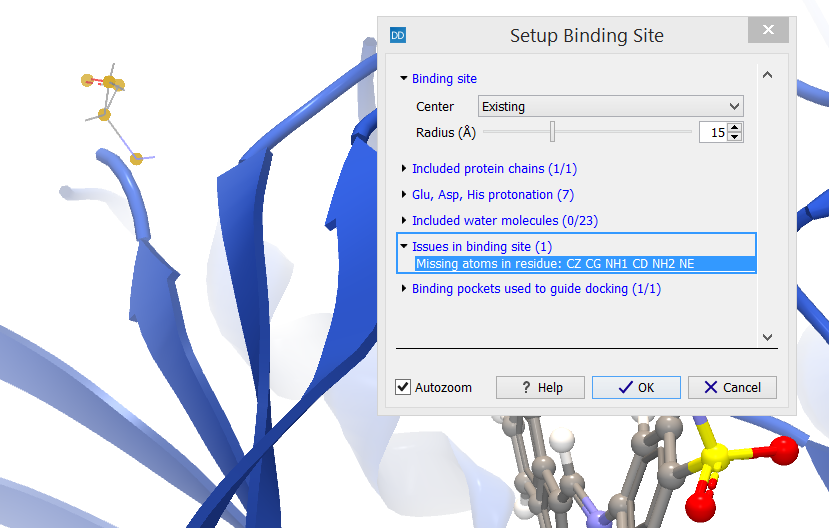
Figure 10.53: The category header indicates the number (X) of the issues listed in the Issues list that relates to atoms or molecules found inside the binding site volume.
The issues found inside the binding site volume are listed. Selecting a row in the list will zoom the view around the implicated atoms and high-light them. If the issues relate to serious deficiencies in the binding pocket, another protein structure should be used to set up the binding site.
Binding pockets used to guide docking (X/Y)
The category header indicates the number of binding pockets that are included in the binding site setup (X) as well as the number of binding pockets that are found inside the binding site volume (Y) (figure 10.54).
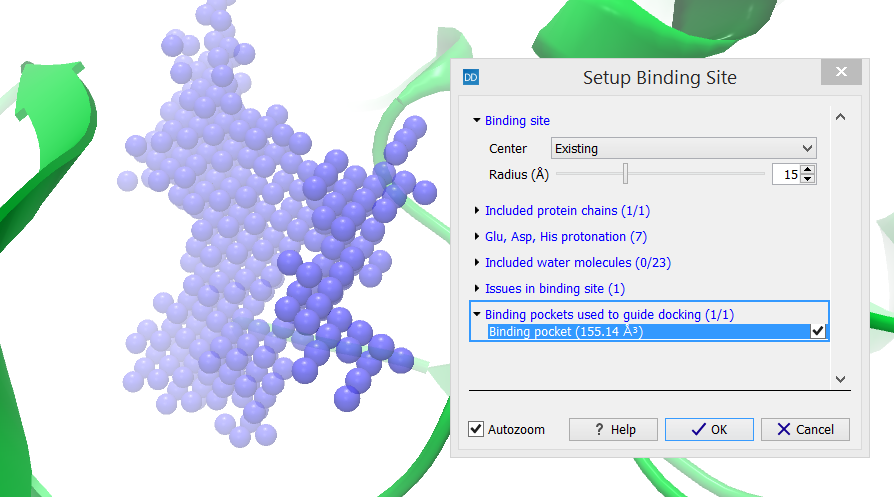
Figure 10.54: The category header indicates the number of binding pockets that are included in the binding site setup (X) as well as the number of binding pockets that are found inside the binding site volume (Y).
Binding pockets are used to guide the docking simulations. Binding modes are enforced to have at least one ligand heavy atom inside a binding pocket included in the setup. It is therefore important that the binding pockets included in the setup actually correspond to where the ligand is expected to bind. All found binding pockets are per default included in the setup, but they can be removed from the setup by un-checking the boxes next to them. The algorithm evaluating the binding pockets is described in Find potential binding pockets.
