Editing atom and bond properties
In some cases, the molecule input contains errors or the automatic assignments are imperfect (see Import Molecules). As the assigned hybridization, hydrogen assignment and bond order is translated into atom types used in the docking simulation, a correct representation is of particular importance for molecules used for docking. If the molecule representation is incorrect, the simulated protein-ligand interactions will also be incorrect. Therefore, molecules can be edited manually, to make the necessary corrections to the representation.For small molecules (ligands), there are two ways to modify the representation. The Ligand Optimizer (The Ligand Optimizer) is a convenient tool for this purpose, but notice that it will sometimes make changes to atom coordinates while doing modifications. Another option, which is available for all categories of molecules, is to edit atom and bond properties directly in the 3D view. The original heavy atom coordinates will always be maintained during these manipulations. To make changes to an atom, right-click the atom, and a list of properties to be used for editing will appear (figure 10.24).
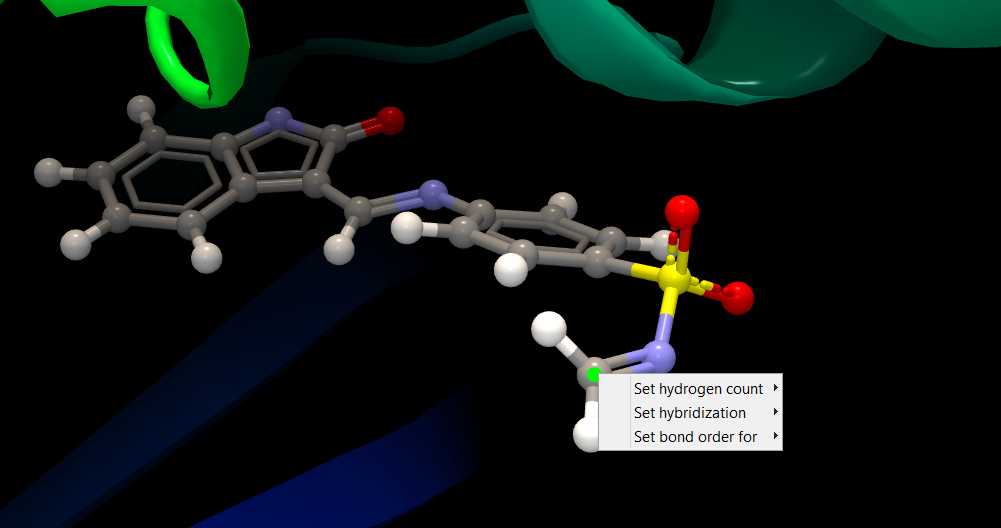
Figure 10.24: To make changes to an atom, right-click the atom, and a list of properties to be used for editing will appear.
- Set hydrogen count will give you a list of the possible number of attached hydrogens to choose from.
- Set hybridization will give you the option of SP1, SP2, and SP3 hybridization. Changing the hybridization will change the position of attached hydrogens, to best adhere to the geometric arrangement around an atom with the requested hybridization. It is not possible to change the atom hybridization for atoms in aromatic rings.
- Set bond order for will give you a list that shows the bonds to neighboring heavy atoms. For each of these, you can pick the order of the bond from a list with the options single, double, triple, and delocalized. It is not possible to change bond order between atoms in aromatic rings.
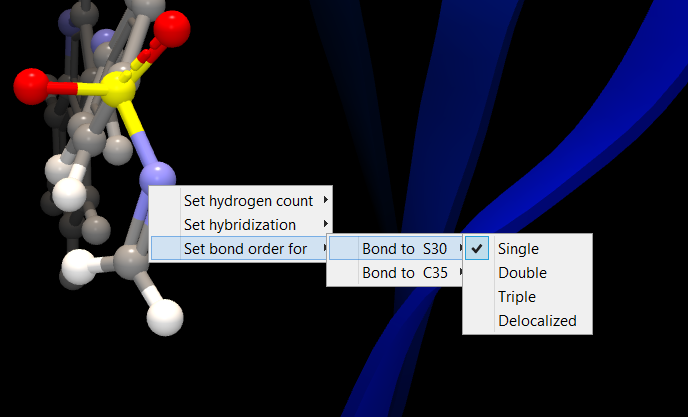
Figure 10.25: To make changes to an atom, right-click the atom, and a list of properties to change will appear.
The Issues view (see Import issues) lists all the concerns that were raised during import of the molecule files. The list is updated whenever changes are made to molecules. The issues listed in the Chemistry category, point out aspects of the molecule representation that seems out of the ordinary. It is therefore a valuable help to have the Issues list in split screen view together with the 3D view, while manually editing molecules. Not having any Chemistry issues connected to a molecule is not a guarantee that the representation is correct, but if there are Chemistry issues, it is relevant to check if they call for changes being made to the molecule.
Please note! If the input file has charges assigned to the atoms, these will also be imported. If the input file has no charges assigned, particular chemical groups (e.g. acids) will be assigned charges during import. However, no charges will or can be assigned at a later stage, and the charges are ignored in the docking simulation. Chemistry issues related to a missing charge can therefore not be amended manually. In such situations it is safe to ignore "Chemistry" issues if the bonding pattern for the atom seems otherwise correct.
Please also note! If you are making changes to a molecule from a Molecule Table, the changes are related to the table, and not the 3D view that was used to specify the changes. It is therefore the Issues list on the Molecule Table that lists the chemistry issues of the molecule, and not the Issues list of the Molecule Project used for visualizing the molecule. Likewise, if you would like to undo a change, the focus should be shifted to the Molecule Table, before clicking the undo button or pressing Ctrl + Z.
