Working with AWS S3 using the Remote Files tab
Browsing data in AWS S3 using the Remote Files tab
The S3 buckets available via the configured AWS Connections are listed under the Remote Files tab, to the right of the Navigation Area tab.
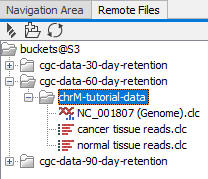
Figure 3.6: AWS S3 buckets you have access to are available under the Remote Files tab.
If more than one AWS Connection is available, for example, if the CLC Workbench is connected to a CLC Server with a valid AWS Connection, a drop-down menu appears above the bucket listing, allowing you to select the AWS Connection to use.
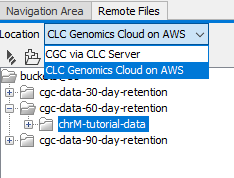
Figure 3.7: This Workbench has a valid AWS Connection and is connected to a CLC Server with a valid AWS Connection. A drop-down menu allows selection of the connection to use. The S3 buckets available using the credentials in that AWS Connection are then listed in the Remote Files tab.
Uploading data to AWS S3 using the Remote Files tab
To upload data from your Navigation Area to AWS S3, right-click on a folder in the Remote Files tab and choose the option Upload to this folder (figure 3.8).
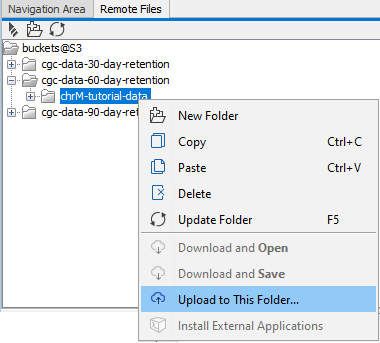
Figure 3.8: To upload data from your Navigation Area to AWS S3, open the Remote Files tab, right-click on the folder you wish to upload data to, and select the option "Upload to This Folder".
Upload is sequential. Information about the data upload is shown in the Processes tab, at the bottom left of the Workbench (figure 3.9).
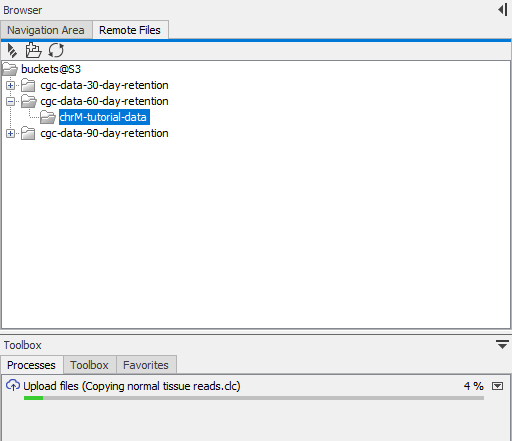
Figure 3.9: After choosing to upload data to S3, the progress of the upload is reported in the Processes tab.
Downloading results via the Remote Files tab
Right-click on an element or elements under the Remote Files tab to download from AWS S3 (figure 3.10). When only a file or files are selected, options to Download and Open and Download and Save will be available. When a folder has been selected, only the Download and Save is available.
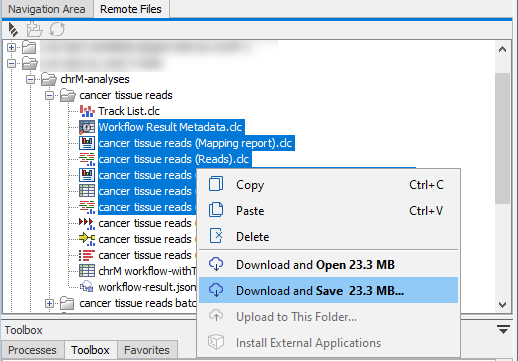
Figure 3.10: Select folders and/or files in the Remote Files tab and right-click to reveal options for downloading that data.
After choosing to download the data, you can choose to download it to a CLC File Location or to another area on your system.
To see all the outputs of a particular job, double-click on a workflow-result.json file. All the results can then be downloaded and opened from that list, or individual elements can be selected and downloaded. The Execution Log is also available from this list (see figure 3.11).
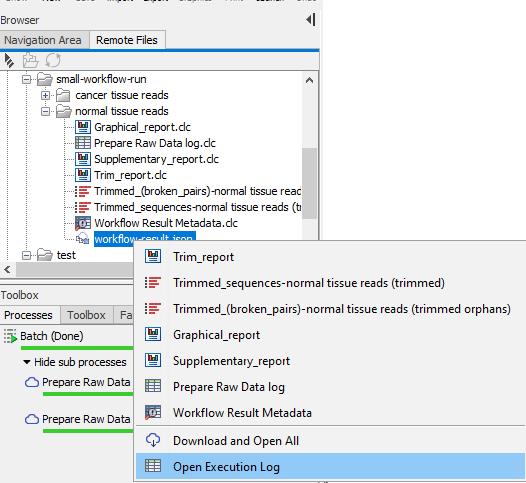
Figure 3.11: Double-click on a workflow-result.json file in the Remote Files tab in the Workbench to reveal a list of all results from that job, as well as the Execution Log. All items can be downloaded and opened from this menu, or individual items can be selected and downloaded.
If the Navigation Tools plugin is installed, bookmarks for items in the Remote Files tab can be made. Double-clicking on bookmarks for individual results files or folders opens the bookmarked items, as standard. Double-clicking on a bookmark for a workflow-result.json file reveals the same list of options as double-clicking on the workflow-result.json file in the Remote Files tab directly. Further details about bookmarks are provided in the Navigation Tools manual at https://resources.qiagenbioinformatics.com/manuals/navigationtools/current/index.php?manual=Introduction.html.
AWS charges for downloaded data from S3. See General information about outputs from cloud analyses for further information about downloading data from AWS S3.
Downloading results using a URL
Pasting an s3 URL into the Workbench Navigation Area will import the files, or folder containing files using Standard Import. File types are automatically detected. Thus, as well as CLC format files being available to view and work further with in the Workbench, other files in formats recognized by Standard Import will be imported as CLC format files, allowing them to be worked with in the Workbench.
Further details about Standard Import are provided at: https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Standard_import.html.
