Whole exome sequencing (WES)
The protein coding part of the human genome accounts for around 1 % of the genome and consists of around 180,000 exons covering an area of 30 megabases (Mb) [Ng et al., 2009]. By targeting sequencing to only the protein coding parts of the genome, exome sequencing is a cost efficient way of generating sequencing data that is believed to harbor the vast majority of the disease-causing mutations [Choi et al., 2009].
Thirteen ready-to-use workflows are available for analysis of whole genome sequencing data (figure 13.1). The concept of the pre-installed ready-to-use workflows is that read data are used as input in one end of the workflow and in the other end of the workflow you get a track based genome browser view and a table with all the identified variants, which may or may not have been subjected to different kinds of filtering and/or annotation.
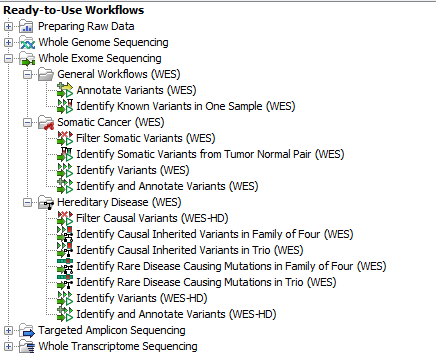
Figure 13.1: The eleven workflows available for analyzing whole exome sequencing data.
In this chapter we will discuss what the individual ready-to-use workflows can be used for and go through step by step how to run the workflows.
Note! Often you will have to prepare data with one of the two Preparing Raw Data workflows described in Preparing Raw Data before you proceed to Analysis of sequencing data (WES).
Subsections
