Trim Primers of Mapped Reads
This tool replaces the previous tools called Trim Primers of Mapped Single Reads and Trim Primers of Mapped Paired End Reads. Both have been moved to the Legacy folder of the Toolbox (see Legacy tools).
The tool Trim Primers of Mapped Reads removes the primer parts of mapped reads (also from RNA-seq mapped reads, except for primers that span intron boundary), as they reflect the primer that was added and not the actual sample. Note that the tool will also remove any insertion located right after the primer part.
The tool can be found in the Toolbox here:
Tools | QIAseq Panel Expert Tools | QIAseq DNA Panel Expert Tools (![]() ) | Trim Primers of Mapped Reads (
) | Trim Primers of Mapped Reads (![]() )
)
In the first dialog (figure 3.20), select a read mapping.
Figure 3.20: Select a read mapping.
In the second dialog (figure 3.21), select the primer annotation track that was provided with the QIAseq DNA Panel.
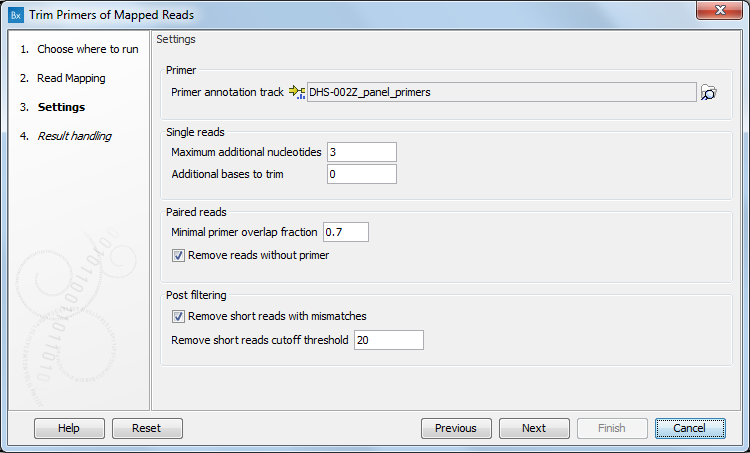
Figure 3.21: Select the primer annotation track specific to the panel, and add the parameters needed to deal with the type of reads you are working with.
In addition, set the following parameters:
- Single reads. This tool works on reads that potentially ends in a primer (rather than starts in a primers). The tool aims to unalign the primer parts of reads that came from that primer. It approximates this by only unaligning reads that end inside of a primer or up to 3 extra bases after the primer, as set by the parameter Maximal additional nucleotides. You can also decide on how many Additional bases to trim right after the primer. This trimming is not done on reads for which dimer artifacts were identified.
- Paired reads. If an aligned read starts within the span of a primer, and if it overlaps the primer with at least 70% (set by default for the Minimal primer overlap fraction option), then it is said to "hit" the primer. For reads "hitting" a primer, the part of the read that overlaps the primer will be unaligned. For reads not "hitting" a primer, the read will either be removed from or retained in the read mapping, depending on the option Remove reads without primer. If the option is checked, the tool will remove reads that do not "hit" a primer.
- Post filtering. After unaligning the primer part of the reads, the tool removes reads where less than 20 aligned bases remain, as set by the parameter Remove short reads cutoff threshold. This feature can be disabled by unchecking the parameter Remove short reads with mismatches.
If one read in a UMI group runs past the primer it overlaps, it means that all reads in that group were not created from that primer. If this happens, then the tool will not unalign any reads in this UMI group.
